Bhardwaj Lab
Quantitative Biology and Data Integration
Institute of Biodynamics and Biocomplexity, Utrecht University

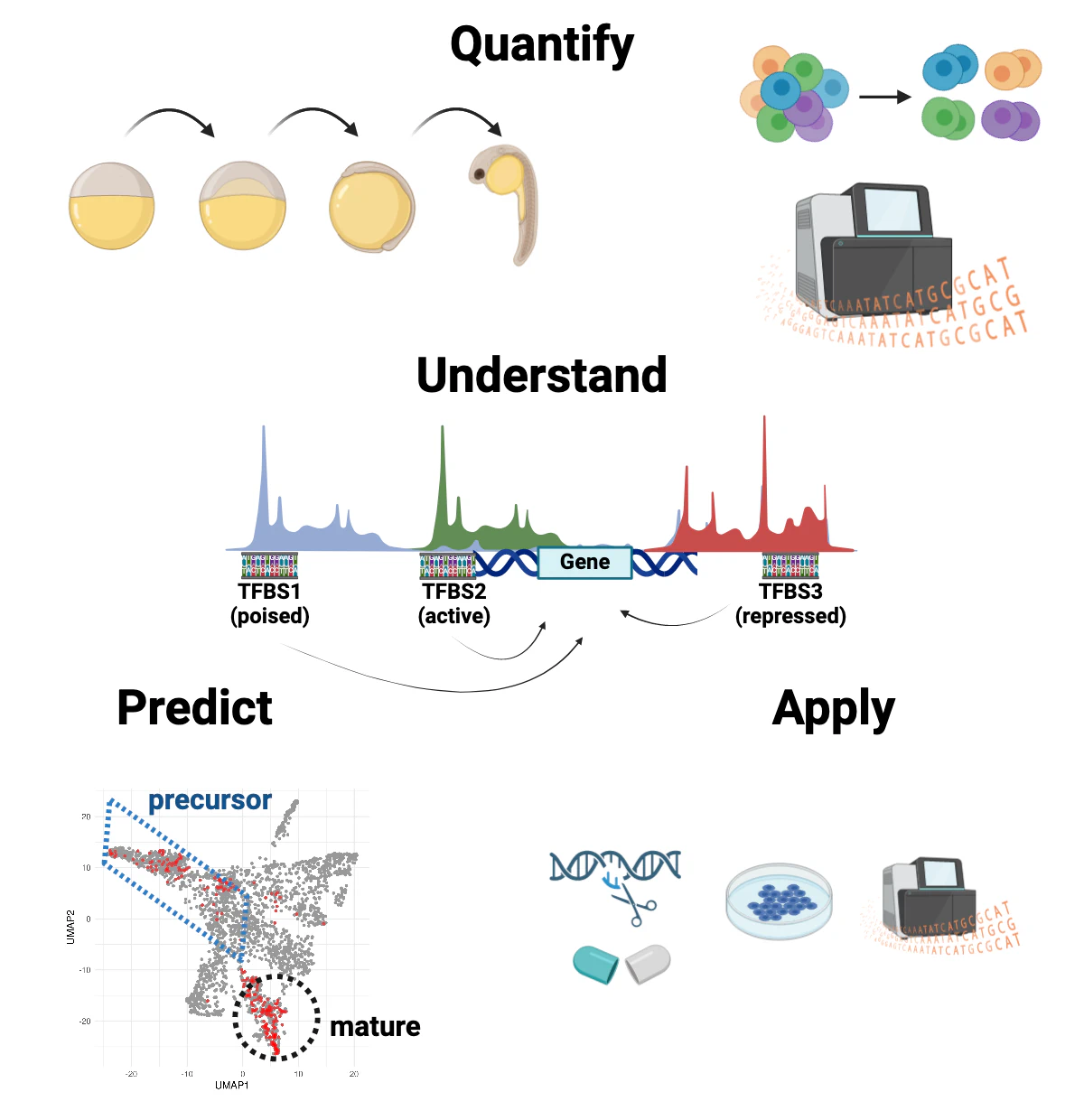
Our mission
The long-term goal of our lab is to transform human medicine by enabling technologies for in-vivo, directed (re)programming of stem cells into specialized cell types.
To acheive this, we need to develop a strong understanding of the fundamental mechanisms that guide the fate decisions for 'naive' stem and pluripotent cells inside our body. During development of an embryo, these cells, which share their genetic code, start to diversify in their 'epigenetic' landscapes. Guided by the regulatory proteins (transcription factors) and their chromatin environment, these cells change their identity in order to perform specific tasks.
By learning the quantitative, dose-response relationship between the epigenetic factors and the decision making process of the cells, we would be able to efficiently reprogram the stem cells into a desired cell type. We can use this understanding to build therapies towards regenerative medicine, healthy aging and cancer.
Our approach
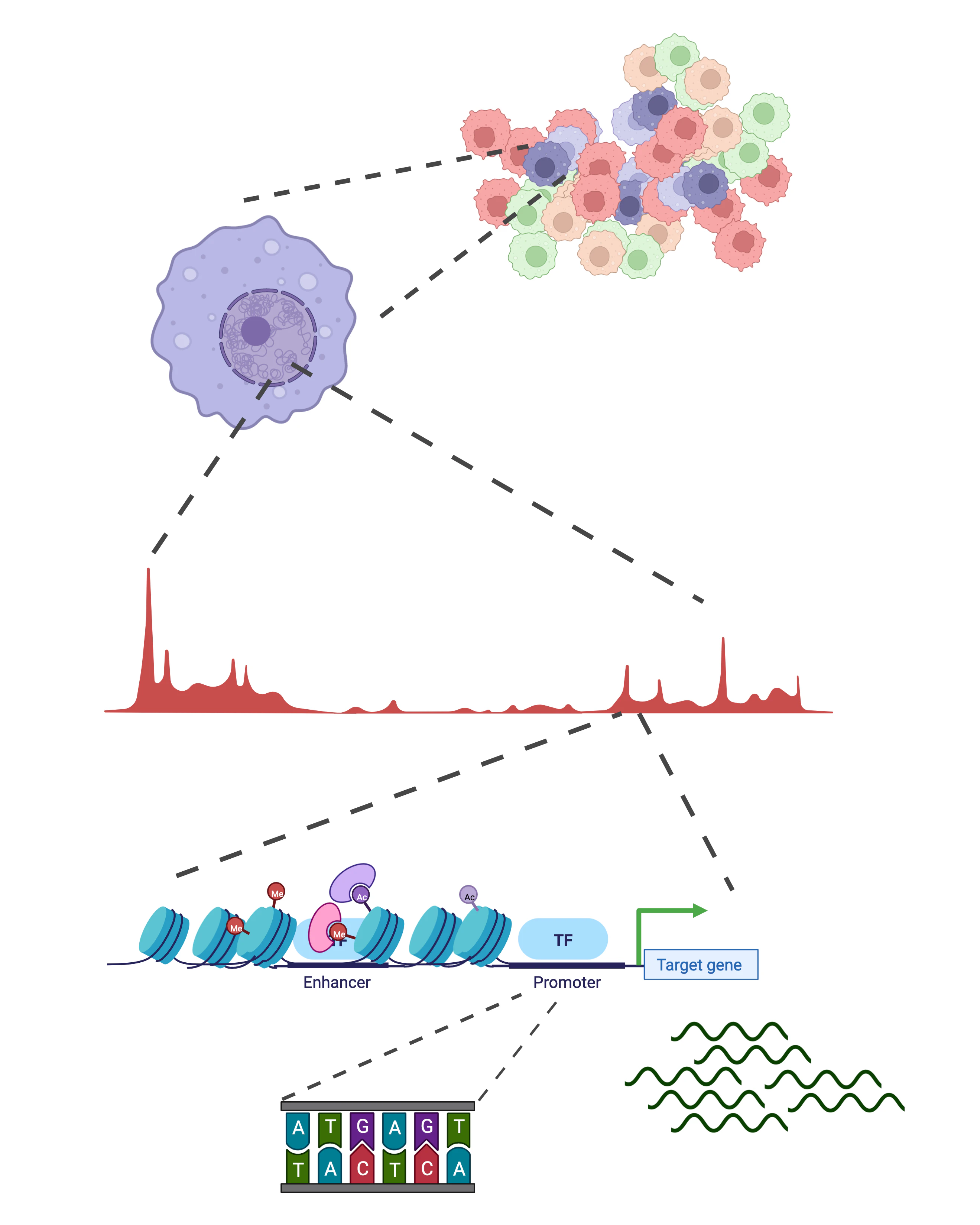
In recent years, several experimental methods have been developed to systematically map the chromatin environment of individual cells, and quantify the gene expression levels of transcription factors. This allows us to build data-driven models that explain the role of these proteins in cellular decision-making.
In our research group, we apply such experimental methods (specifically, single-cell and single-molecule genomics technologies) to understand cellular decision-making. These methods produce large scale data, typically measuring thousands of genes over thousands of cells, molecules, or samples. We use these datasets to develop statistical and machine learning models that can explain the relationship between the regulators (transcription factors, chromatin state), and a cell's current or future state.
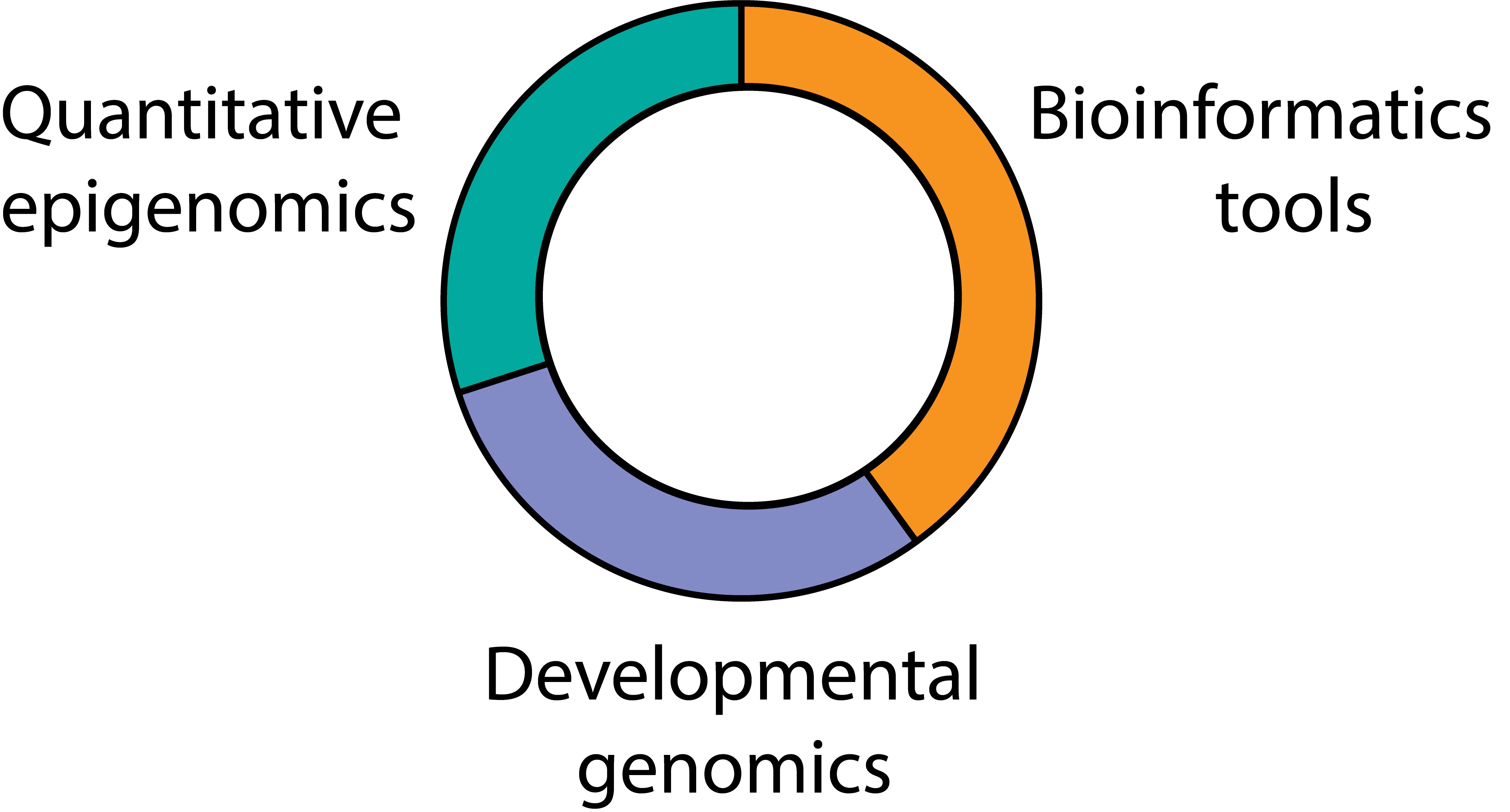
Our Projects
We apply a data-driven biology approach to our projects. Half of our work is focussed on extracting new insights from quantitative epigenomics and multi-omics data of developing cells, with the aim to learn new mechanistic models of cell identity and function. The rest half of our work is focussed on building new bioinformatic methods and tools to analyse the data from such technologies, and making them available to biologists.
News
Latest developments from our lab

We just released a new preprint describing our single cell multiomics analysis of histone modifications and gene expression of zebrafish embryos

vivek
Stay Up to Date
Subscribe to our lab updates/open positions


